Information Processing in the Cell Nucleus
The human genome presents a fascinating puzzle: it contains around 20,000 protein-coding genes, however, nearly one million regulatory elements control when and how these genes are expressed. This disparity raises intriguing questions about how such a limited number of genes can lead to the vast diversity of cell types and functions in complex organisms. At the Institute of Biological and Chemical Systems – Biological Information Processing (IBCS-BIP) at Karlsruhe Institute of Technology (KIT), we attempt to answer these questions by studying the processing of genomic information in the cell nucleus. Our research profits from the use of zebrafish as a model for how the genetic code controls embryonic development and a panel of inbred medaka fish lines as a model for how genetic diversity encodes phenotype differences in a natural population. We also dissect the mechanistic underpinnings of how gene regulatory elements control the read-out of specific genes, and venture into creating synthetic DNA-sequence-programmable materials that mimic the natural control of gene read-out. By understanding how living systems store and process information, we hope to discover innovative approaches towards bio-inspired materials and technologies for information processing, ultimately benefiting fields such as biotechnology and medicine.
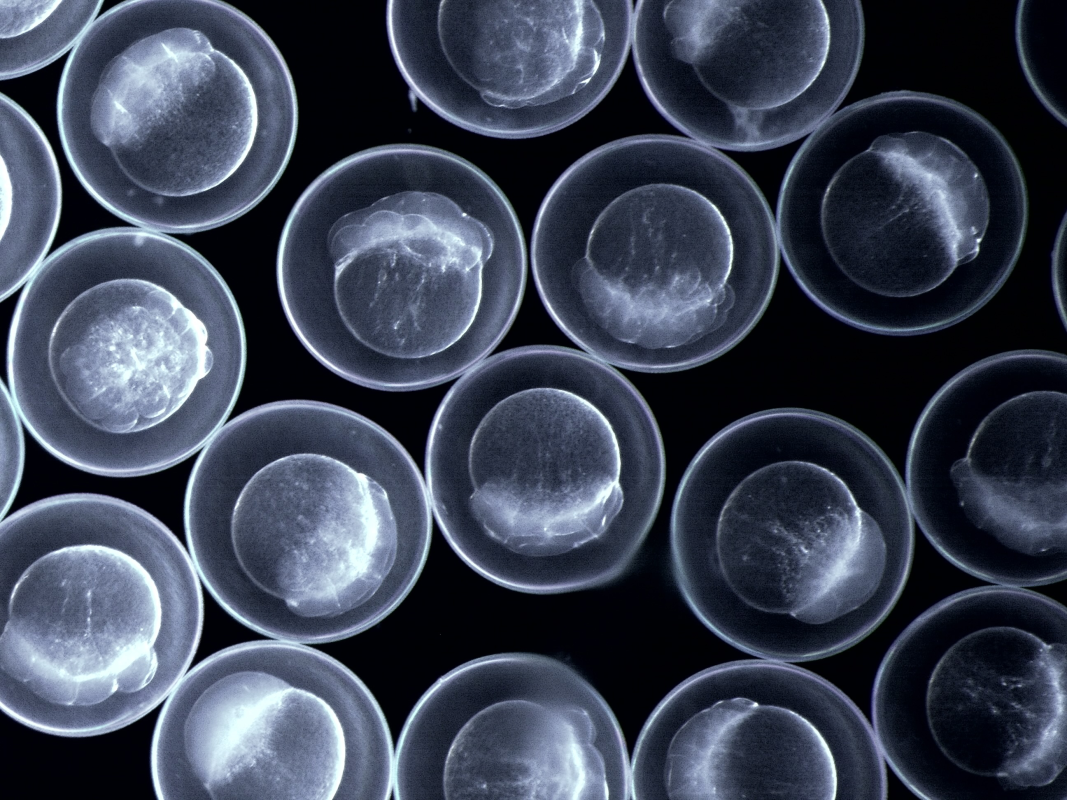
Decoding the Zebrafish Genome
“The DANIO-CODE consortium has identified over 140,000 cis-regulatory elements that control gene transcription throughout zebrafish development.”
A significant achievement in understanding nuclear information storage has been our contribution to the DANIO-CODE consortium, which established a centralized repository for zebrafish developmental genomic data. This collaborative effort has resulted in a comprehensive resource containing 1,802 sets of genomic data, including both unpublished and re-analyzed published information [1]. This repository has proven invaluable for improving existing annotations and demonstrating their practical value in experimental design.
Key findings from this work include:
- Identification of over 140,000 cis-regulatory elements that control gene transcription throughout zebrafish development.
- Mapping of unique chromatin and spatial topologies between regulatory elements active during early zygotic genome activation and those active later in organogenesis.
- Alignment of regulatory elements and epigenomic landscapes between zebrafish and mouse, revealing functional relationships beyond simple sequence similarity.
These insights provide a deeper understanding of how genetic information is organized and accessed during vertebrate development, offering potential applications in both basic research and biomedical studies.
Exploring Genotype-Phenotype Relationships
Building upon the genomic insights gained from zebrafish studies, we have established a panel of medaka inbred strains to further investigate the genetic basis of natural trait variation in vertebrates. The establishment of a panel of 80 medaka inbred strains from a wild medaka population, a task requiring more than 6 years and 2000 crosses, has resulted in each line serving as a “snap shot” of genetic variability in that wild population. Given that the genome sequence is available for each line, genome-wide association studies allow us to explore the genetic underpinnings of various complex traits.
Initial studies focused on resting heart rate—a well-established predictor of overall mortality. Using wide-field microscopy, we quantified embryonic heart rate variations between strains. Subsequent genotype-phenotype association studies identified several genomic regions associated with heart rate frequency, including genes linked to heart development and organ maintenance.
This medaka inbred panel represents a powerful resource for future studies on the genetic basis of various complex traits and diseases, potentially leading to new insight in developmental biology and personalized medicine.
Nuclear Structure and Function
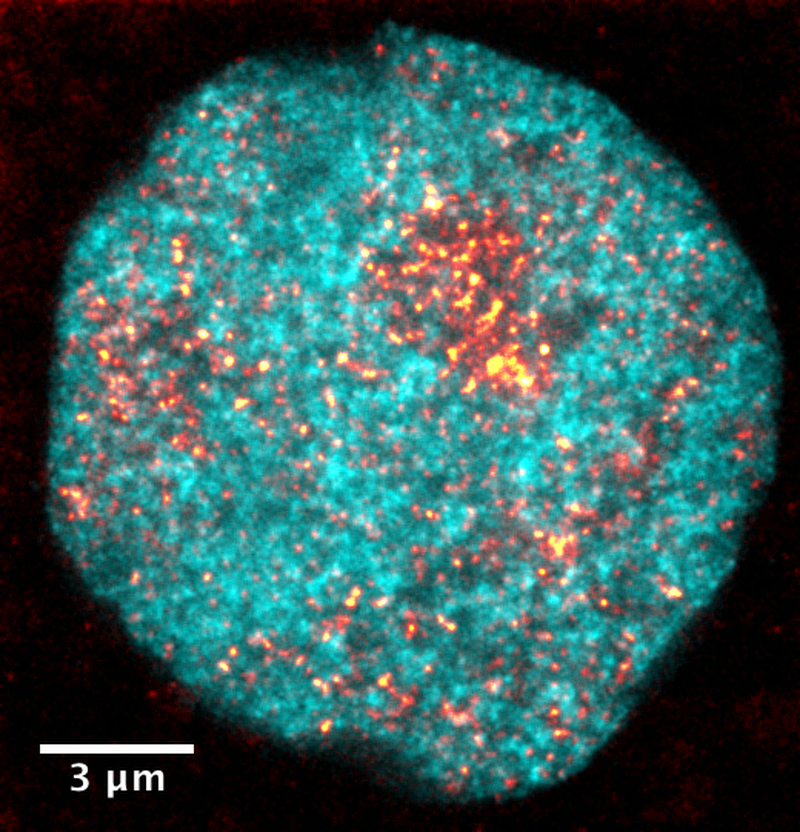
DNA in blue, active polymerase in orange.
Recent advances in imaging technologies have led to new insight into the structure and physicochemical properties of the cell nucleus, revealing their contribution to the organization and controlled read-out of genetic information.
“Euchromatin is organized into sub-100-nm nanodomains as constitutive units, which can only be seen by super-resolution microscopy.”
Euchromatin Organization:
Combining the latest improvements of 3D-STED super-resolution microscopy with novel fluorescence stains, we have scrutinized euchromatin organization in zebrafish embryos as a model for embryonic pluripotency [2,3]:
- Euchromatin is organized into sub-100-nm nanodomains as constitutive units, which can only be visualized by super-resolution microscopy.
- Active transcription sites stabilize these nanodomains into a finely dispersed configuration unique to embryonic pluripotency.
- The finely dispersed state can be theoretically represented as a microphase-separated polymer suspension. Genomic regions harboring ongoing transcription act to stabilize this organization similar to macromolecular amphiphiles.
These observations underscore the unique, fine-structured organization of euchromatin during pluripotency in embryonic development. The modelling of this structure as a microphase-separated polymer material further hints at how such an organization could be artificially recreated.
We have developed novel, purely optical techniques, with which we can study the mechanical properties, or “rheology”, of specific genomic regions. These purely optical techniques can be applied in living cells without inflicting damage during measurements. By creating localized temperature gradients using infrared illumination, we reveal [4]:
- Substantial intra-nuclear chromatin displacements in response to the temperature gradients, allowing chromatin rheology while maintaining overall nuclear organization.
- Distinct mechanical properties are apparent for different nuclear compartments containing different epigenetic states of chromatin.
- A high resistance to deformation in both dense heterochromatin and loosely packed euchromatin compared to a lower resistance to deformation in medium-dense chromatin.
Thus, our newly developed biophotonics technologies reveal previously inaccessible rheological aspects of functional nuclear organization in a non-invasive fashion, thereby enabling exciting opportunities in stem cell biology and drug screening.
Dynamic Gene Regulation:
To extract additional information from the latest cutting-edge microscopy techniques, we have applied artificial intelligence (AI) and machine learning methods to imaging data from zebrafish embryos and mouse embryonic stem cells (mESC). Building on these approaches, we have gained more insight into gene-regulatory processes:
- The discovery that stem cell-specific clusters of RNA polymerase II form on regulatory chromatin regions, which provide surfaces for localized liquid-phase condensation. [5]
- The development of a protocol to embed quality control with AI-based denoising enabled accelerated imaging of post-translational modifications and three-dimensional shapes of RNA polymerase II clusters in live zebrafish embryos. [6]
- The demonstration that pluripotency-specific genes visit RNA polymerase II clusters in stereotyped events, which last only a few seconds. These events were extracted from the accelerated live-sample data and additionally validated against pseudo-time inference from fixed-cell data.
These findings reveal how transient visits of genes to macromolecular clusters which last only a few seconds represent a specialized mode of transcription control unique to pluripotency. Furthermore, our work demonstrates that the fundamental material process of surface condensation underlies this special type of transcription control.
Synthetic DNA Nanostructures:
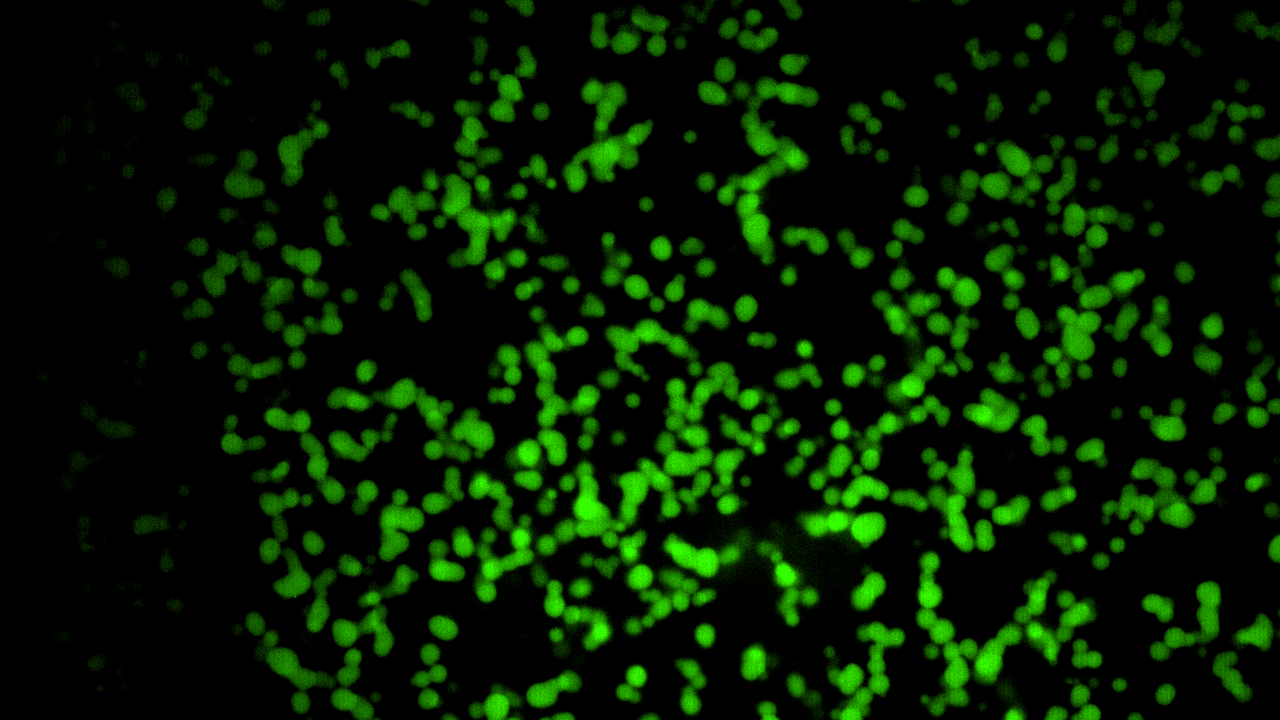
“We have constructed synthetic, DNA-sequence-programmable nanostructures that mimic key aspects of nuclear information processing.”
Building on the insight we have gained regarding the material and rheological characteristics of natural nuclear structures, we have constructed synthetic, DNA-sequence-programmable nanostructures that mimic key aspects of nuclear information processing. This work follows two major directions:
- Design of DNA-nanostructures that assemble into hydrogels. The programmability via DNA sequence allows both the fine-tuning of mechanical properties as well as the functional functionalization of these hydrogels. [7]
- Creation of DNA-nanomotifs that undergo liquid-phase condensation similar to stem-cell-specific RNA polymerase II clusters. These nanomotifs can be selectively programmed to target specific synthetic DNA strands and can be equipped with amphiphilic properties. Both aspects mimic key properties of the transcriptional clusters observed in stem cells. [8]
These biomimetic approaches allow us to reproduce key aspects of nuclear information processing, such as chromatin rheology and stem-cell-specific transcriptional clusters. We can thereby open new avenues for constructing synthetic nanomaterials with DNA-sequence-programmable properties.
Outlook and Implications
The research on information processing in the cell nucleus at KIT not only advances our understanding of fundamental biological mechanisms but also holds transformative potential for applications in biotechnology, medicine, and synthetic biology. By unraveling the complex interplay between genetic information storage and regulatory control, we are paving the way for innovative solutions that could redefine how we approach diagnostics, therapeutic interventions, and the development of bio-inspired materials. As we continue to explore these intricate systems, our work promises to contribute to both scientific knowledge and practical advancements in various fields.
References
[1] Multiomic atlas with functional stratification and developmental dynamics of zebrafish cis-regulatory elements. Damir Baranasic ... Ferenc Müller, 2022. Nature Genetics, 54(9), pp.1382-1394.
[2] Transcription organizes euchromatin via microphase separation. Lennart Hilbert, Yuko Sato, Ksenia Kuznetsova, Tommaso Bianucci, Hiroshi Kimura, Frank Jülicher, Alf Honigmann, Vasily Zaburdaev, Nadine L Vastenhouw, 2021. Nature Communications, 12(1), p.1360.
[3] The hierarchical packing of euchromatin domains can be described as multiplicative cascades. Amra Noa, Hui-Shun Kuan, Vera Aschmann, Vasily Zaburdaev, Lennart Hilbert, 2021. PLOS Computational Biology, 17, p.e1008974.
[4] Probe-free optical chromatin deformation and measurement of differential mechanical properties in the nucleus. Benjamin Seelbinder, Susan Wagner, Manavi Jain, Elena Erben, Sergei Klykov, Iliya Dimitrov Stoev, Venkat Raghavan Krishnaswamy, Moritz Kreysing, 2024. eLife, 13, p.e76421.
[5] RNA polymerase II clusters form in line with surface condensation on regulatory chromatin. Agnieszka Pancholi, Tim Klingberg, Weichun Zhang, Roshan Prizak, Irina Mamontova, Amra Noa, Marcel Sobucki, Andrei Yu Kobitski, Gerd Ulrich Nienhaus, Vasily Zaburdaev, Lennart Hilbert, 2021. Molecular Systems Biology, 17(8), p.e10272.
[6] Deep-learning microscopy image reconstruction with quality control reveals second-scale rearrangements in RNA polymerase II clusters. Hamideh Hajiabadi, Irina Mamontova, Roshan Prizak, Agnieszka Pancholi, Anne Koziolek, Lennart Hilbert, 2022. PNAS Nexus, 1(3), p. pgac065.
[7] Quantitative Characterization of RCA‐Based DNA Hydrogels–Towards Rational Materials Design. Svenja A Moench, Phillip Lemke, Julia Weisser, Iliya D Stoev, Kersten S Rabe, Carmen M Domínguez, Christof M Niemeyer, 2024. Chemistry–A European Journal, 30(53), p.e202401788.
[8] Amphiphiles formed from synthetic DNA-nanomotifs mimic the stepwise dispersal of transcriptional clusters in the cell nucleus. Xenia Tschurikow, Aaron Gadzekpo, Mai P Tran, Rakesh Chatterjee, Marcel Sobucki, Vasily Zaburdaev, Kerstin Göpfrich, Lennart Hilbert, 2023. Nano Letters, 23(17), p.7815-7824.

.jpg)