Zebrafish Endocrinology and Metabolism
Research interest
The Dickmeis laboratory explores the zebrafish as a model organism to study the interplay between hormonal signaling and metabolism in development and disease. A main focus is placed on the temporal dynamics of metabolism and endocrine regulation.
To this end, we develop biosensors to measure hormonal and metabolic signaling activity in the living animal, targeting e.g. glucocorticoid hormones, glucose or reactive oxygen species. We also employ systems biology approaches (transcriptomics, proteomics, metabolomics) to understand the temporal dynamics of metabolism and their regulation by endocrine and metabolic signaling, both during development and regeneration and across the day-night cycle. In chemical in vivo screens with zebrafish embryos, we apply tools and concepts derived from our studies to evaluate toxicity of chemicals and to identify novel lead compounds for drug development.
Below are highlighted three current projects:
 The aim of the EU funded international PrecisionTox project is to better protect the health of people and the environment by establishing New Approach Methodologies (NAMs) for chemical safety testing using a mix of genomics, metabolomics, evolutionary theory, quantitative genetics, data science, toxicology, and law. The consortium uses human cell lines and a diverse suite of well-established biomedical model organisms — fruit flies, water fleas, round worms, and embryos of frogs and zebrafish — as well as artificial intelligence (AI) approaches to uncover molecular toxicity pathways shared across the animal kingdom.
The aim of the EU funded international PrecisionTox project is to better protect the health of people and the environment by establishing New Approach Methodologies (NAMs) for chemical safety testing using a mix of genomics, metabolomics, evolutionary theory, quantitative genetics, data science, toxicology, and law. The consortium uses human cell lines and a diverse suite of well-established biomedical model organisms — fruit flies, water fleas, round worms, and embryos of frogs and zebrafish — as well as artificial intelligence (AI) approaches to uncover molecular toxicity pathways shared across the animal kingdom.
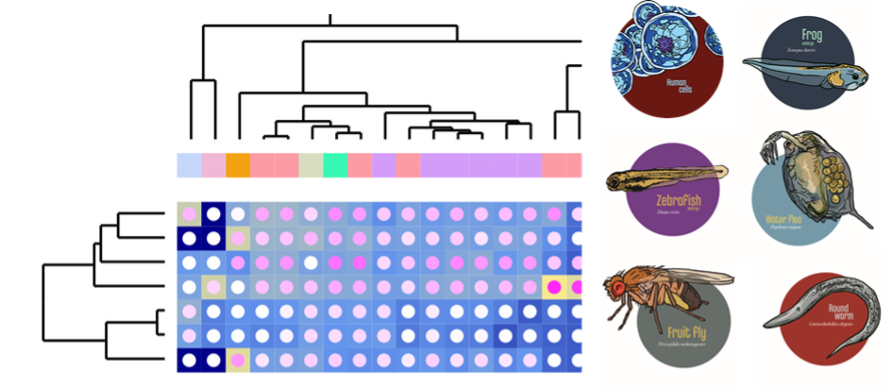
Within this project, we contribute our expertise on zebrafish embryos and examine chemical compounds specifically for their metabolic and endocrine disrupting activities. In vivo assays targeting metabolic regulation and hormonal signaling anchor the information obtained from OMICS data generated by the consortium to specific compound-induced phenotypes, generating mechanistic knowledge that can be used to better understand, predict and treat toxic compound effects. We closely collaborate with the group led by Carsten Weiss, which covers the human cell culture part of the project.
![]() A second EU funded project, Toxbox, aims to develop a flexible, all-in-one platform for comprehensive chemical toxicity testing, which also provides advanced data integration and reliable computer models for better risk assessment. This platform will include a zebrafish module which allows to record biochemical, microscopic and behavioural readouts from zebrafish embryos treated with chemicals. We collaborate with Ralf Mikut’s group at IAI/KIT, also a partner in this consortium, to develop machine learning approaches for data evaluation from zebrafish embryo assays.
A second EU funded project, Toxbox, aims to develop a flexible, all-in-one platform for comprehensive chemical toxicity testing, which also provides advanced data integration and reliable computer models for better risk assessment. This platform will include a zebrafish module which allows to record biochemical, microscopic and behavioural readouts from zebrafish embryos treated with chemicals. We collaborate with Ralf Mikut’s group at IAI/KIT, also a partner in this consortium, to develop machine learning approaches for data evaluation from zebrafish embryo assays.
Role of reactive oxygen species (ROS) in brain regeneration
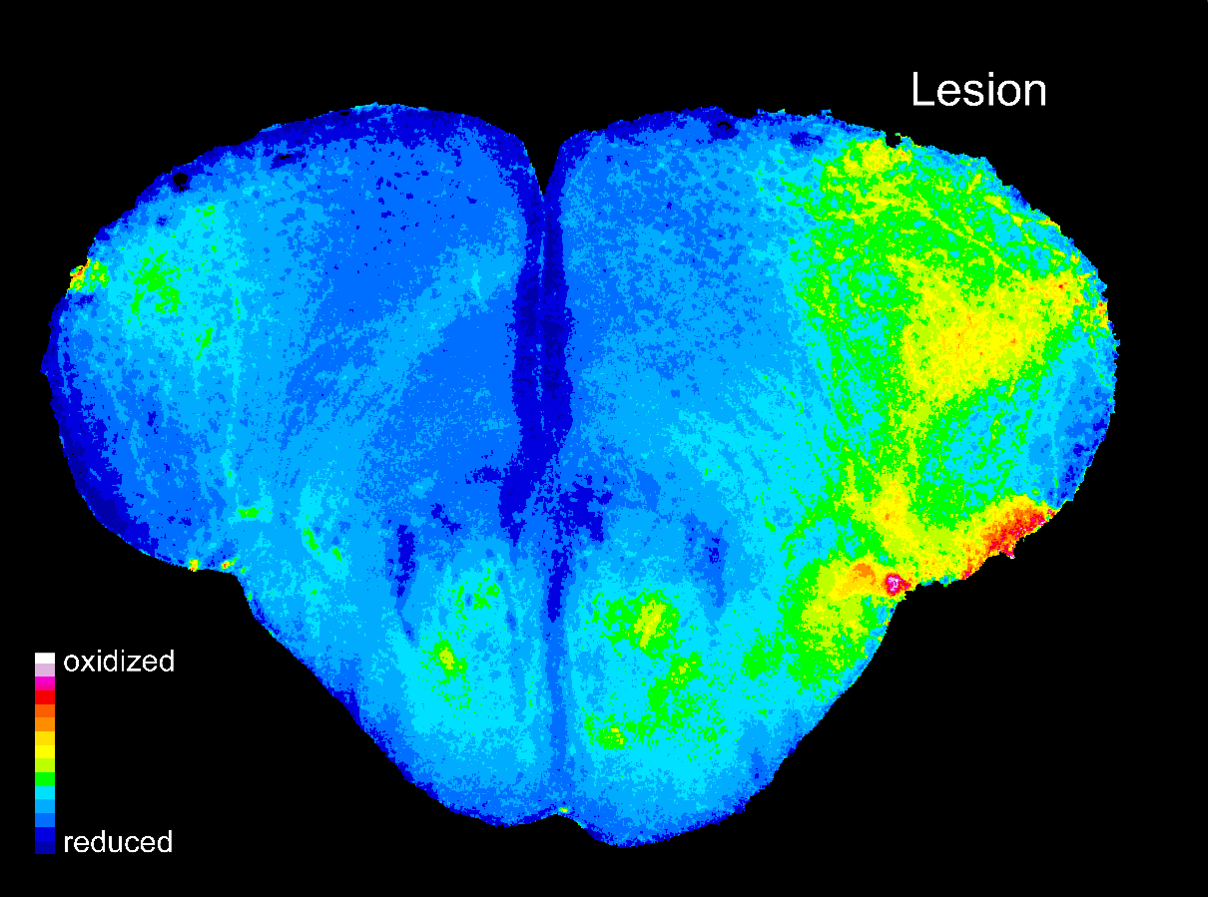
In contrast to mammals including humans, zebrafish have the remarkable capability to regenerate damaged neuronal tissue within a few weeks. In our project, we apply molecular tools to visualize metabolites and ROS during the process of brain regeneration after physical injury. In this way, we hope to obtain insight into potential functions of altered metabolism and redox states during the injury response and subsequent regenerative processes.
Dr. Thomas Dickmeis
- principal investigator
- Group: Dickmeis
- Room: CN
- Phone: +49 721 608-26564
- thomas dickmeis ∂does-not-exist.kit edu
- Hermana-von-Helmholtz-Platz 1
76344 Eggenstein-Leopoldshafen
Publications
Hayot, G.; Lloyd, G. R.; Diwan, G. D.; Keith, N.; Smoot, S. R.; Cramer von Clausbruch, C. A.; Kaufman, T. C.; Barnard, M.; Tindall, A. J.; Glaholt, S. P.; Massei, R.; Martínez, R.; Strähle, U.; Orsini, L.; Russell, R. B.; Tennessen, J. M.; Scholz, S.; Shaw, J. R.; Freedman, J. H.; Colbourne, J. K.; Weiss, C.; Dickmeis, T.
2025. Environmental Science & Technology, 59 (48), 25634–25648. doi:10.1021/acs.est.5c10177
Martínez, R.; González-Sánchez, J. C.; Sampani, S. I.; Scholz, S.; Escher, B. I.; Henneberger, L.; Huchthausen, J.; Whelan, M.; Dickmeis, T.; Weiss, C.; Colbourne, J. K.; Freedman, J. H.
2025. Toxicological Sciences, 208 (2), 317–329. doi:10.1093/toxsci/kfaf126
Siauciuinate, R.; Etard, C.; Koehler, A.; Pisanty, O.; Otto, M.; Dickmeis, T.; Kassel, O.; Gothilf, Y.; Foulkes, N. S.; Vallone, D.
2025. Laboratory Animals, 59 (6), 664–675. doi:10.1177/00236772251351087
Yan, J.; Takamiya, M.; Zhang, D.; Pace, G.; Rastegar, S.; Wang, H.; Schoch, S.; Köberle, B.; Hartwig, A.; Dickmeis, T.; Weiss, C.
2025. Environment International, 197, Article no: 109349. doi:10.1016/j.envint.2025.109349
Lautenschlager, T.; Friederich, N.; Sitcheu, A. J. Y.; Nau, K.; Hayot, G.; Dickmeis, T.; Mikut, R.
2025. arxiv. doi:10.48550/arXiv.2510.07853
Hayot, G.; Massei, R.; Lloyd, G.; Keith, N.; Diwan, G.; Martínez López, R.; Barnard, M.; Cramer von Clausbruch, C. A.; Grasse, N.; Smoot, S.; Escher, B.; Tennessen, J.; Tindall, A.; Oliver, B.; Shaw, J.; Scholz, S.; Freedman, J.; Strähle, U.; Colbourne, J. K.; Weiss, C.; Dickmeis, T.
2024. Naunyn-Schmiedeberg’s archives of pharmacology, 397 (Suppl. 1), S17–69. doi:10.1007/s00210-024-02974-3
Squarisi, I. S.; Ribeiro, V. P.; Ribeiro, A. B.; Souza, L. T. M. de; Junqueira, M. de M.; Oliveira, K. M. de; Hayot, G.; Dickmeis, T.; Bastos, J. K.; Veneziani, R. C. S.; Ambrósio, S. R.; Tavares, D. C.
2024. Pharmaceuticals, 17 (10), Art.-Nr.: 1340. doi:10.3390/ph17101340
Hayot, G.; Marcato, D.; Cramer von Clausbruch, C. A.; Pace, G.; Strähle, U.; Colbourne, J. K.; Pylatiuk, C.; Peravali, R.; Weiss, C.; Scholz, S.; Dickmeis, T.
2024. Journal of visualized experiments : JoVE, 203, Art.-Nr.: e66153. doi:10.3791/66153
Weiss, C.; Dickmeis, T.; Hayot, G.; Peravali, R.; Cramer von Clausbruch, C.; Pace, G.
2023. Toxicology Letters, 383, 33–42. doi:10.1016/j.toxlet.2023.05.004
Cramer von Clausbruch, C.; Weiss, C.; Dickmeis, T.; Peravali, R.; Strähle, U.; Colbourne, J.; Hayot, G.; Scholz, S.; Lopez, R. M.
2023. Naunyn-Schmiedeberg’s Archives of Pharmacology, 396 (S1), S61–62. doi:10.1007/s00210-023-02397-6
Dickmeis, T.; Hayot, G.; Clausbruch, C. C. von; Martinez Lopez, R.; Scholz, S.; Colbourne, J.; Strähle, U.; Weiss, C.; Peravali, R.
2023. Naunyn-Schmiedeberg’s Archives of Pharmacology, 396 (S1), S61
Hansjosten, I.; Takamiya, M.; Rapp, J.; Reiner, L.; Fritsch-Decker, S.; Mattern, D.; Andraschko, S.; Anders, C.; Pace, G.; Dickmeis, T.; Peravali, R.; Rastegar, S.; Strähle, U.; Hsiao, I.-L.; Gilliland, D.; Ojea-Jimenez, I.; Ambrose, S. V. Y.; Belinga-Desaunay-Nault, M.-F. A.; Khan, A. O.; Lynch, I.; Valsami-Jones, E.; Diabaté, S.; Weiss, C.
2022. Environmental science / Nano, 91 (1), 375–392. doi:10.1039/d1en00299f
Breus, O.; Dickmeis, T.
2020. Biological chemistry, 402 (3), 363–378. doi:10.1515/hsz-2020-0269
Weger, M.; Weger, B. D.; Schink, A.; Takamiya, M.; Stegmaier, J.; Gobet, C.; Parisi, A.; Kobitski, A. Y.; Mertes, J.; Krone, N.; Strähle, U.; Nienhaus, G. U.; Mikut, R.; Gachon, F.; Gut, P.; Dickmeis, T.
2020. eLife, 9, Art. Nr. e57068. doi:10.7554/eLife.57068
Takamiya, M.; Stegmaier, J.; Kobitski, A. Y.; Schott, B.; Weger, B. D.; Margariti, D.; Cereceda Delgado, A. R.; Gourain, V.; Scherr, T.; Yang, L.; Sorge, S.; Otte, J. C.; Hartmann, V.; Wezel, J. van; Stotzka, R.; Reinhard, T.; Schlunck, G.; Dickmeis, T.; Rastegar, S.; Mikut, R.; Nienhaus, G. U.; Strähle, U.
2020. PLoS Genetics, 16 (6), Art. Nr.: e1008774. doi:10.1371/journal.pgen.1008774
Adhikari, S. S.; Zhao, L.; Dickmeis, T.; Korvink, J. G.; Badilita, V.
2019. The analyst, 144 (24), 7192–7199. doi:10.1039/C9AN01634A
Dickmeis, T.; Feng, Y.; Mione, M. C.; Ninov, N.; Santoro, M.; Spaink, H. P.; Gut, P.
2019. Frontiers in cell and developmental biology, 7, 15. doi:10.3389/fcell.2019.00015
Neukum, A.; Bartschat, A.; Breitwieser, H.; Strähle, U.; Dickmeis, T.; Pylatiuk, C.
2019. Zebrafish, 16 (3), 326–328. doi:10.1089/zeb.2019.1728
Leidner, A.; Bauer, J.; Ebrahimi Khonachah, M.; Takamiya, M.; Strähle, U.; Dickmeis, T.; Rabe, K. S.; Niemeyer, C. M.
2019. Biomaterials, 190-191, 76–85. doi:10.1016/j.biomaterials.2018.10.035
Weger, M.; Weger, B. D.; Görling, B.; Poschet, G.; Yildiz, M.; Hell, R.; Luy, B.; Akcay, T.; Güran, T.; Dickmeis, T.; Müller, F.; Krone, N.
2018. EBioMedicine, 36, 376–389. doi:10.1016/j.ebiom.2018.09.024
Breitwieser, H.; Dickmeis, T.; Vogt, M.; Ferg, M.; Pylatiuk, C.
2018. SLAS TECHNOLOGY: Translating Life Sciences Innovation, 23 (2), 128–133. doi:10.1177/2472630317745780
Weger, M.; Diotel, N.; Dorsemans, A. C.; Dickmeis, T.; Weger, B. D.
2017. Developmental biology, 431 (2), 111–123. doi:10.1016/j.ydbio.2017.09.012
Tsachaki, M.; Meyer, A.; Weger, B.; Kratschmar, D. V.; Tokarz, J.; Adamski, J.; Belting, H.-G.; Affolter, M.; Dickmeis, T.; Odermatt, A.
2017. The journal of endocrinology, 232 (2), 323–335. doi:10.1530/JOE-16-0495
Geisler, R.; Köhler, A.; Dickmeis, T.; Strähle, U.
2017. EMBO reports, 18 (1), 1–2. doi:10.15252/embr.201643561
Weger, B. D.; Weger, M.; Görling, B.; Schink, A.; Gobet, C.; Keime, C.; Poschet, G.; Jost, B.; Krone, N.; Hell, R.; Gachon, F.; Luy, B.; Dickmeis, T.
2016. PLoS Genetics, 12 (12), e1006512. doi:10.1371/journal.pgen.1006512
Schutera, M.; Dickmeis, T.; Mione, M.; Peravali, R.; Marcato, D.; Reischl, M.; Mikut, R.; Pylatiuk, C.
2016. Bioengineered, 7 (4), 261–265. doi:10.1080/21655979.2016.1197710
Shahid, M.; Takamiya, M.; Stegmaier, J.; Middel, V.; Gradl, M.; Klüver, N.; Mikut, R.; Dickmeis, T.; Scholz, S.; Rastegar, S.; Yang, L.; Strähle, U.
2016. Scientific Reports, 6, Art.Nr. 23768. doi:10.1038/srep23768
Takamiya, M.; Weger, B. D.; Schindler, S.; Beil, T.; Yang, L.; Armant, O.; Ferg, M.; Schlunck, G.; Reinhard, T.; Dickmeis, T.; Rastegar, S.; Strähle, U.; Leung, Y. F.
2015. PLoS ONE, 10 (2), e0117645. doi:10.1371/journal.pone.0117645
Ferg, M.; Armant, O.; Yang, L.; Dickmeis, T.; Rastegar, S.; Strähle, U.
2014. Briefings in functional genomics, 13, 131–143. doi:10.1093/bfgp/elt044
Dickmeis, T.; Weger, B. D.; Weger, M.
2013. Molecular and Cellular Endocrinology, 380, 2–15. doi:10.1016/j.mce.2013.05.012
Tixier, V.; Bataille, L.; Etard, C.; Jagla, T.; Weger, M.; DaPonte, J. P.; Strähle, U.; Dickmeis, T.; Jagla, K.
2013. Proceedings of the National Academy of Sciences of the United States of America, 110, 18982–18987. doi:10.1073/pnas.1301262110
Mikut, R.; Dickmeis, T.; Driever, W.; Geurts, P.; Hamprecht, F. A.; Kausler, B. X.; Ledesma-Carbayo, M. J.; Maree, R.; Mikula, K.; Pantazis, P.; Ronneberger, O.; Santos, A.; Stotzka, R.; Strähle, U.; Peyrieras, N.
2013. Zebrafish, 10, 401–421. doi:10.1089/zeb.2013.0886
Weger, B. D.; Weger, M.; Jung, N.; Lederer, C.; Bräse, S.; Dickmeis, T.
2013. Journal of Visualized Experiments, 79, e50439/1–9. doi:10.3791/50439
Weger, M.; Weger, B. D.; Diotel, N.; Rastegar, S.; Hirota, T.; Kay, S. A.; Strähle, U.; Dickmeis, T.
2013. Developmental Biology, 380, 259–273. doi:10.1016/j.ydbio.2013.04.035
Weger, B. D.; Weger, M.; Nusser, M.; Brenner-Weiss, G.; Dickmeis, T.
2012. ACS Chemical Biology, 7, 1178–1183. doi:10.1021/cb3000474
Weger, B. D.; Sahinbas, M.; Otto, G. W.; Mracek, P.; Armant, O.; Dolle, D.; Lahiri, K.; Vallone, D.; Ettwiller, L.; Geisler, R.; Foulkes, N. S.; Dickmeis, T.
2011. PLoS One, 6 (2), e17080/1–15. doi:10.1371/journal.pone.0017080
Dickmeis, T.; Foulkes, N. S.
2011. Molecular and Cellular Endocrinology, 331, 11–22. doi:10.1016/j.mce.2010.09.001
Dickmeis, T.; Sahinbas, M.; Weger, B. D.
2009. Asia-Pacific Journal of Endocrinology, 1 (1), 75–86
Dickmeis, T.
2009. The journal of endocrinology, 200 (1), 3–22. doi:10.1677/JOE-08-0415
Rastegar, S.; Hess, I.; Dickmeis, T.; Nicod, J. C.; Ertzer, R.; Hadzhiev, Y.; Thies, W. G.; Scherer, G.; Strähle, U.
2008. Developmental Biology, 318, 366–77. doi:10.1016/j.ydbio.2008.03.034
Pezeron, G.; Lambert, G.; Dickmeis, T.; Strähle, U.; Rosa, F. M.; Mourrain, P.
2008. PLoS One, 3 (1), e1434
Plessy, C.; Dickmeis, T.; Chalmel, F.; Strähle, U.
2005. Trends in Genetics, 21, 207–10. doi:10.1016/j.tig.2005.02.006
Dickmeis, T.; Müller, F.
2005. Briefings in Functional Genomics and Proteomics, 3, 332–50
Read More
Award for best short talk to Gaëlle Hayot at the 9th German Pharm-Tox Summit in Munich!
Gaëlle Hayot (Dickmeis lab) received an award for the “Best Short Talk in the Field of Toxicology” by the German Gesellschaft für Toxikologie for her work on “Systematic acquisition of toxicity data in non-sentient models across animal phylogeny: implications for read-across and estimation of toxicity in humans”, presented at the 9th German Pharm-Tox Summit in Munich. Congratulations!
Best paper award ESN
Our collaborative work involving the screening centre and three IBCS-BIP groups (Dickmeis-, Rastegar-, Weiss-lab) received the Environmental Science: Nano best paper award 2022! Within the EU FP7 project NanoMILE, we joined forces with scientists from different disciplines to study the impact of specifically synthesized and well characterized nanomaterials on a variety of cell lines and zebrafish embryos. Ten years after the project was launched, we finally published our detailed analysis on the importance of surface properties of nanomaterials for detrimental effects on zebrafish development. Such highly interdisciplinary work across countries is indeed required to support the safe development of nanomaterials and truly inspiring, but also challenging, for all the researchers involved at different career stages ranging from bachelor students, PhD and post-doctoral students up to senior scientists (https://pubs.rsc.org/en/content/articlelanding/2023/en/d3en90021e).
Award for the best presentation at the Annual Consortium Meeting of the European Project Precision Toxicology
At the Annual Consortium Meeting of the European Project Precision Toxicology held in Barcelona in June 2022, Dr. Gaëlle Hayot (Dickmeis lab) and PhD Student Christina Cramer von Clausbruch (Weiss lab) won not only two nice coffee mugs but also the award for the best presentation showcasing their work on zebrafish and human cells as models to improve chemical safety. Congratulations!
PrecisionTox Podcast
Podcast by Thomas Dickmeis on the zebrafish model: from ecotoxicology to biomedical research (https://precisiontox.org/voxlab/).
Precision Toxicology consortium aims to protect human health from effects of harmful chemicals
A major research project to shape regulation and policy on chemical safety without the use of animal testing has been launched with the aid of €19.3M funding from the European Commission.
Interview about the project in Wiley Analytical Science: https://analyticalscience.wiley.com/content/news-do/auswirkungen-von-chemikalien-auf-die-menschliche-gesundheit

