Medaka fish, a vertebrate genetic model system
 |
Fig. 1: Wild male medaka. Medaka is a freshwater teleost from East Asia and well established model system (Aida, 1921). Small body size, short generation time, high fecundity, compact genome and high tolerance to inbreeding render medaka well suited for many studies. |
The MIKK panel of inbred medaka lines, a population genetic resource
Individuals of wild populations are genetically polymorphic. Thus, even though they share the same set of genes, their genomes contain many sequence polymorphisms that affect phenotypic traits, for example morphology, physiology and behaviour. Often several polymorphisms, mostly SNPs, act on a given trait and lead to subtle quantitative phenotypic variations. A classical example is craniofacial variation of human individuals (Fig. 2). The repertoire of genetic variations in a population and their effects on a given trait are studied by genome wide association studies (GWAS) that use statistical analysis to elucidate phenotype-genotype associations. Functional tests are required to validate these statistical associations. Therefore, suitable animal models are needed.


Fig. 2: Individual facial variation in humans and wild Kiyosu medaka (6 billion others, Auer)
A vertebrate animal model for GWAS should allow phenotypic and genetic analysis of a large number of individuals (> 1000) to achieve statistical significance of association. Inbred lines are important to permit experimental replication using the same set of genetic variants (i.e. genetic background). Medaka (Oryzias latipes) is a well established and fecund genetic vertebrate model and fulfills these criteria as it has a high tolerance to inbreeding from the wild (Hyodo-Taguchi and Egami, 1985). Furthermore medaka wild catches can easily be obtained from their natural habitat in Japan (Spivakov et al., 2014). In collaboration with Jochen Wittbrodt (COS, HD Germany), Ewan Birney (EBI, UK), and Kiyoshi Naruse (NBRP, Japan) we have established a medaka population genetic resource to be used as a vertebrate animal model for GWAS. Importantly in addition to association studies, the inbred panel allows testing of associated SNPs by in vivo functional assays (genome editing).
 |
wild polymorphic population
|
Fig. 3: Starting with random crosses of individuals from the wild population a panel of inbred strains was established by inbreeding for several generations. Each inbred line harbors a different set of genetic variants.
At the medaka facility of IBCS-BIP we have established a panel of 80 near-isogenic medaka inbred lines from a carefully selected wild population without prior genetic domestication of founder animals (Fig. 3). We used single full-sibling-pair inbreeding for 9 generations to breed the panel known as the Medaka Inbred Kiyosu-Karlsruhe (MIKK) panel (Fitzgerald et al., 2022). Whole genome sequence analysis showed that more than 75% of the strains are more than 80% homozygous, a value which is comparable to the Drosophila genetic reference panel. The entire panel captures more than 25 million SNPs of the wild population.
In addition to the high number of inbred lines, the MIKK panel is unique among vertebrate inbred panels as it has been derived from a wild population. It is well known that the domestication of wild animals often has severe consequences for the genetic diversity of a population. In the MIKK panel, genetic adaptation to husbandry conditions prior to inbreeding did not occur.
Complex genetic traits are quantitative traits and therefore require quantitative phenotyping assays. Assays must be scalable to permit phenotyping of large numbers of individuals to reach statistical significance. We have developed several assays to phenotype specific traits.
In collaboration with Tilo Baumbach, IPS/KIT we have developed an X-ray micro-CT imaging pipeline including automated segmentation for quantitative morphometric analysis of embryonic and adult medaka fish (Weinhardt et al., 2018). This detailed morphometric analysis aims to identify genetic variants regulating morphometric variation (Fig. 4).
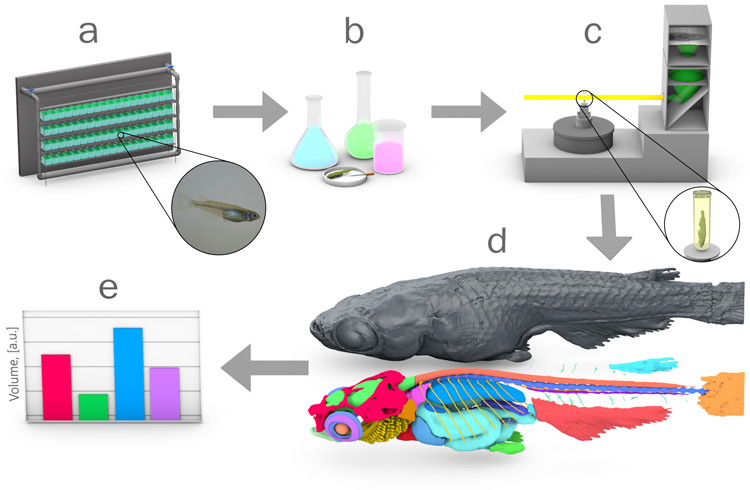
Fig. 4: Pipeline for quantitative 3D morphometric analysis. (a) fish husbandry; (b) fixation, staining with contrast agent and embedding; (c) X-ray micro-CT at the synchrotron radiation facility; (d) 3D reconstruction and manual segmentation of the reference atlas; (e) automatic segmentations and morphometric analysis. (Weinhardt et al., 2018).
In addition to this morphological analysis we are using organ function assays, such as analysis of the resting heart rate. We are interested in the genetic control of the resting heart rate as it is a well-established predictor of overall mortality. An association study in humans revealed an association of heart rate genetic variants and all cause mortality, such that 5bpm increased heart rate results in a decreased life span of about 2.5 years, underscoring the importance of resting heart rate as a key risk factor (Eppinga et al., 2016).
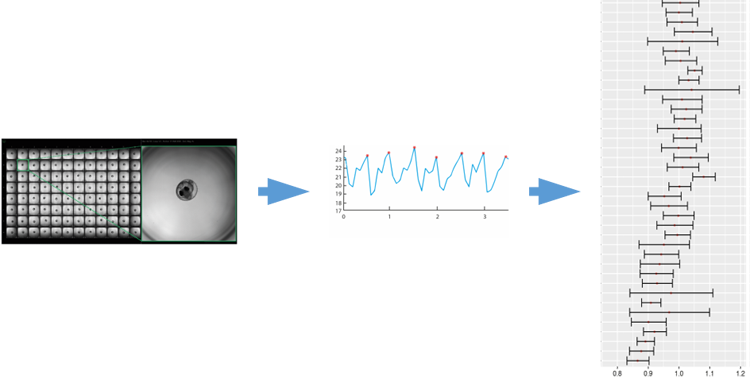
Fig. 5: Quantitative assay to determine resting heart rate. Native resting heart rate of embryos is extracted from short videos by automated image analysis. Right panel shows variance of normalized heart rate in some strains of the inbred panel (Gierten et al., 2020).
The Kiyosu panel allows to study the underlying genetics of this key trait and to functionally test associated genetic variants. We quantify native embryonic heart rate with wide field microscopy in a 96 well format by extracting the frequency of heartbeat from a short image sequence (Fig. 5; Gierten et al., 2020).
In a pilot study utilizing highly inbred strains of medaka (Oryzias latipes, and Oryzias sakaizumii), automated quantification of embryonic heart rates as core metric revealed phenotype variability across five isogenic inbred strains. Experimental validation of the top 12 candidate genes in loss-of-function models revealed their causal and distinct impact on heart rate, development, ventricle size, and arhythmia (Gierten et al, 2023). In an analogous genetic approach, genetic variants regulating the sommitic clock have been identified (Seleit et al., 2024).
We also use the MIKK strains to test human SNPs for functional relevance, which is not possible in humans (Hammouda et al., 2021). It is therefore possible to identify and evaluate causative genetic variants causing individual trait variation.
Comparison of data sets from different phenotyping assays across the panel will allow to search for pleiotropic effects of genetic variants. Cross species comparison by comparative genomics will enhance the association power of GWAS data. Associated genetic variants will be tested by CRISPR/Cas genome editing in specific strains of the medaka inbred panel. We have developped further assays to phenotype physiological and behavioural traits with the aim to provide a more complete phenotypic description of the individual strains and to establish a data base of functionally relevant genetic variants of the medaka Kiyosu population.
References:
Aida, T., (1921). On the Inheritance of Color in a Fresh-Water Fish, APLOCHEILUS LATIPES Temmick and Schlegel, with Special Reference to Sex-Linked Inheritance. Genetics 6, 554–573.
Eppinga, R. N., Hagemeijer, Y., Burgess, S., Hinds, D. A., Stefansson, K., Gudbjartsson, D. F., van Veldhuisen, D. J., Munroe, P. B., Verweij, N. and van der Harst, P., (2016). Identification of genomic loci associated with resting heart rate and shared genetic predictors with all-cause mortality. Nat Genet 48, 1557–1563.
Fitzgerald, T., Brettell, I., Leger, A., Wolf, N., Kusminski, N., Monahan, J., Barton, C., Herder, C., Aadepu, N., Gierten, J., Becker, C., Hammouda, O., Hasel, E., Lischik, C., Lust, K., Sokolova, N., Suzuki, R., Tsingos, E., Tavhelidse, T., Thumberger, T., Watson, P., Welz, B., Khouja, N., Naruse, K., Birney, E., Wittbrodt, J., & Loosli, F. (2022). The Medaka Inbred Kiyosu-Karlsruhe (MIKK) panel. Genome Biology, 23(1), 59.
Gierten, J., Pylatiuk, C., Hammouda, O., Schock, C., Stegmaier, J., Wittbrodt, J., Gehrig, J., & Loosli, F. (2020). Automated high-throughput heartbeat quantification in medaka and zebrafish embryos under physiological conditions. Scientific Reports, 10(1), 2046.
Gierten, J., Welz, B., Fitzgerald, T., Thumberger, T., Hummel, O., Leger, A., Weber, P., Hassel, D., Hübner, N., Birney, E., & Wittbrodt, J. (2023). Natural genetic variation quantitatively regulates heart rate and dimension. bioRxiv, 2023.09.01.555906.
Hammouda, O., Wu, M., Kaul, V., Gierten, J., Thumberger, T., & Wittbrodt, J. (2021). In vivo identification and validation of novel potential predictors for human cardiovascular diseases. PLoS ONE, 16(12), e0261572.
Hyodo-Taguchi, Y. and Egami, N. (1985). Establishment of Inbred Strains of the Medaka Oryzias latipes and the Usefulness of the Strains for Biomedical Research. Zool. Sci. 2, 305–316.
Seleit, A., Brettell, I., Fitzgerald, T., Vibe, C., Loosli, F., Wittbrodt, J., Naruse, K., Birney, E., & Aulehla, A. (2024). Modular control of vertebrate axis segmentation in time and space. The EMBO Journal, 1–24.
Spivakov, M., Auer, T. O., Peravali, R., Dunham, I., Dolle, D., Fujiyama, A., Toyoda, A., Aizu, T., Minakuchi, Y., Loosli, F., et al. (2014). Genomic and phenotypic characterization of a wild medaka population: towards the establishment of an isogenic population genetic resource in fish. G3 (Bethesda) 4, 433–445.
Weinhardt, V., Shkarin, R., Wernet, T., Wittbrodt, J., Baumbach, T., & Loosli, F. (2018). Quantitative morphometric analysis of adult teleost fish by X-ray computed tomography. Scientific Reports, 8(1), 16531.
Dr. Felix Loosli
- PI
- Group: Loosli, Medaka Facility
- Room:
CN 319 - Phone: +49 721 608-28743
- felix loosli ∂does-not-exist.kit edu
- www.itg.kit.edu/loosli.php
- Hermann-von-Helmholtz-Platz 1
76344 Eggenstein-Leopoldshafen
Publications
Naruse, K.; Loosli, F.; Ansai, S.; Birney, E.; Wittbrodt, J.
2026. Trends in Genetics. doi:10.1016/j.tig.2025.12.005
Welz, B.; Pierotti, S.; Fitzgerald, T.; Thumberger, T.; Suzuki, R.; Watson, P.; Fuss, J.; Cordeiro da Trindade, T.; Defranoux, F.; Ferreira, M.; Naruse, K.; Loosli, F.; Gierten, J.; Wittbrodt, J.; Birney, E.
2026. Cell Genomics, Art.Nr: 101164. doi:10.1016/j.xgen.2026.101164
Pierotti, S.; Brettell, I.; Fitzgerald, T.; Herder, C.; Aadepu, N.; Pylatiuk, C.; Wittbrodt, J.; Birney, E.; Loosli, F.
2025. Bioinformatics, 41 (6), Article no: btaf342. doi:10.1093/bioinformatics/btaf342
Pierotti, S.; Brettell, I.; Fitzgerald, T.; Herder, C.; Aadepu, N.; Pylatiuk, C.; Wittbrodt, J.; Birney, E.; Loosli, F.
2024. Cold Spring Harbor Laboratory. doi:10.1101/2024.10.18.618696
Seleit, A.; Brettell, I.; Fitzgerald, T.; Vibe, C.; Loosli, F.; Wittbrodt, J.; Naruse, K.; Birney, E.; Aulehla, A.
2024. The EMBO Journal, 43 (18), 4068–4091. doi:10.1038/s44318-024-00186-2
Sousa-Ortega, A.; Vázquez-Marín, J.; Sanabria-Reinoso, E.; Corbacho, J.; Polvillo, R.; Campoy-López, A.; Buono, L.; Loosli, F.; Almuedo-Castillo, M.; Martínez-Morales, J. R.
2023. Nature Communications, 14, Art.-Nr.: 2804. doi:10.1038/s41467-023-38482-w
Lucon-Xiccato, T.; Loosli, F.; Conti, F.; Foulkes, N. S.; Bertolucci, C.
2022. Scientific Reports, 12 (1), Art.Nr. 10926. doi:10.1038/s41598-022-14978-1
Leger, A.; Brettell, I.; Monahan, J.; Barton, C.; Wolf, N.; Kusminski, N.; Herder, C.; Aadepu, N.; Becker, C.; Gierten, J.; Hammouda, O. T.; Hasel, E.; Lischik, C.; Lust, K.; Sokolova, N.; Suzuki, R.; Tavhelidse, T.; Thumberger, T.; Tsingos, E.; Watson, P.; Welz, B.; Naruse, K.; Loosli, F.; Wittbrodt, J.; Birney, E.; Fitzgerald, T.
2022. Genome biology, 23 (1), Article no: 58. doi:10.1186/s13059-022-02602-4
Fitzgerald, T.; Brettell, I.; Leger, A.; Wolf, N.; Kusminski, N.; Monahan, J.; Barton, C.; Herder, C.; Aadepu, N.; Gierten, J.; Becker, C.; Hammouda, O. T.; Hasel, E.; Lischik, C.; Lust, K.; Sokolova, N.; Suzuki, R.; Tsingos, E.; Tavhelidse, T.; Thumberger, T.; Watson, P.; Welz, B.; Khouja, N.; Naruse, K.; Birney, E.; Wittbrodt, J.; Loosli, F.
2022. Genome biology, 23 (1), Article no: 59. doi:10.1186/s13059-022-02623-z
Lucon-Xiccato, T.; Montalbano, G.; Frigato, E.; Loosli, F.; Foulkes, N. S.; Bertolucci, C.
2022. Hormones and Behavior, 145, Article no: 105244. doi:10.1016/j.yhbeh.2022.105244
Agostini, C.; Bühler, A.; Antico Calderone, A.; Aadepu, N.; Herder, C.; Loosli, F.; Carl, M.
2022. Frontiers in Cell and Developmental Biology, 10, Art.-Nr.: 1005776. doi:10.3389/fcell.2022.1005776
Bruch, R.; Scheikl, P. M.; Mikut, R.; Loosli, F.; Reischl, M.
2021. SLAS technology, 26 (4), 367–376. doi:10.1177/2472630320977454
López-Olmeda, J. F.; Zhao, H.; Reischl, M.; Pylatiuk, C.; Lucon-Xiccato, T.; Loosli, F.; Foulkes, N. S.
2021. iScience, 24 (7), Art. Nr.: 102784. doi:10.1016/j.isci.2021.102784
Soergel, H.; Loosli, F.; Muhle-Goll, C.
2021. Metabolites, 11 (11), Art.-Nr.: 744. doi:10.3390/metabo11110744
Gierten, J.; Pylatiuk, C.; Hammouda, O. T.; Schock, C.; Stegmaier, J.; Wittbrodt, J.; Gehrig, J.; Loosli, F.
2020. Scientific reports, 10 (1), Article no: 2046. doi:10.1038/s41598-020-58563-w
Lucon-Xiccato, T.; Conti, F.; Loosli, F.; Foulkes, N. S.; Bertolucci, C.
2020. Biology, 9 (11), Article: 389. doi:10.3390/biology9110389
Pylatiuk, C.; Zhao, H.; Gursky, E.; Reischl, M.; Peravali, R.; Foulkes, N.; Loosli, F.
2019. SLAS technology, 24 (4), 394–398. doi:10.1177/2472630319841412
Weinhardt, V.; Shkarin, R.; Wernet, T.; Wittbrodt, J.; Baumbach, T.; Loosli, F.
2018. Scientific reports, 8 (1), Art. Nr.: 16531. doi:10.1038/s41598-018-34848-z
Gierten, J.; Loosli, F.; Gehrig, J.; Pylatiuk, C.; Fitzgerald, T.; Birney, E.; Wittbrodt, J.; Gorenflo, M.
2018. The thoracic and cardiovascular surgeon, 66 (S02). doi:10.1055/s-0038-1628326
Gierten, J.; Loosli, F.; Gehrig, J.; Pylatiuk, C.; Fitzgerald, T.; Birney, E.; Wittbrodt, J.; Gorenflo, M.
2018. 50th Annual Meeting of the German Society for Pediatric Cardiology (DGPK 2018), Leipzig, Germany, February 17–20, 2018
Lindsey, B. W.; Douek, A. M.; Loosli, F.; Kaslin, J.
2018. Frontiers in neuroscience, 11. doi:10.3389/fnins.2017.00750
Pereiro, L.; Loosli, F.; Fernández, J.; Härtel, S.; Wittbrodt, J.; Concha, M. L.
2017. Developmental dynamics, 246 (11), 812–826. doi:10.1002/dvdy.24496
Aghaallaei, N.; Gruhl, F.; Schaefer, C. Q.; Wernet, T.; Weinhardt, V.; Centanin, L.; Loosli, F.; Baumbach, T.; Wittbrodt, J.
2016. Development <Cambridge>, 143 (19), 3470–3480. doi:10.1242/dev.134098
Zhang, P.; Elabd, S.; Hammer, S.; Solozobova, V.; Yan, H.; Bartel, F.; Inoue, S.; Henrich, T.; Wittbrodt, J.; Loosli, F.; Davidson, G.; Blattner, C.
2015. Oncogene, 34, 5729–5738. doi:10.1038/onc.2015.21
Kirchmaier, S.; Naruse, K.; Wittbrodt, J.; Loosli, F.
2015. Genetics, 199 (4), 905–918. doi:10.1534/genetics.114.173849
Spivakov, M.; Auer, T. O.; Peravali, R.; Dunham, I.; Dolle, D.; Fujiyama, A.; Toyoda, A.; Aizu, T.; Minakuchi, Y.; Loosli, F.; Naruse, K.; Birney, E.; Wittbrodt, J.
2014. G3: Genes, genomes, genetics, 4 (3), 433–445. doi:10.1534/g3.113.008722
Zaucker, A.; Bodur, T.; Roest Crollius, H.; Hadzhiev, Y.; Gehrig, J.; Loosli, F.; Watson, C.; Müller, F.
2014. Zebrafish, 11 (6), 509–517. doi:10.1089/zeb.2014.0984
Cuesta, I. H.; Lahiri, K.; Lopez-Olmeda, J. F.; Loosli, F.; Foulkes, N. S.; Vallone, D.
2014. Chronobiology international, 31 (4), 468–478. doi:10.3109/07420528.2013.856316
Loosli, F.
2013. Small GTPases, 4, 242–246. doi:10.4161/sgtp.27061
Herder, C.; Swiercz, J. M.; Müller, C.; Peravali, R.; Quiring, R.; Offermanns, S.; Wittbrodt, J.; Loosli, F.
2013. Development, 140, 2787–2797. doi:10.1242/dev.096487
Ravi, V.; Bhatia, S.; Gautier, P.; Loosli, F.; Tay, B. H.; Tay, A.; Murdoch, E.; Coutinho, P.; Heyningen, V.; Brenner, S.; Venkatesh, B.; Kleinjan, D. A.
2013. PLoS Genetics, 9, e1003177/1–24. doi:10.1371/journal.pgen.1003177
d’Alencon, C. A.; Pena, O. A.; Wittmann, C.; Gallardo, V. E.; Jones, R. A.; Loosli, F.; Liebel, U.; Grabher, C.; Allende, M. L.
2010. BMC Biology, 151/1-16, 8. doi:10.1186/1741-7007-8-151
Brown, K. E.; Keller, P. J.; Ramialison, M.; Rembold, M.; Stelzer, E. H. K.; Loosli, F.; Wittbrodt, J.
2010. Developmental Biology, 339, 14–25. doi:10.1016/j.ydbio.2009.12.003
Signore, I. A.; Guerrero, N.; Loosli, F.; Colombo, A.; Villalon, A.; Wittbrodt, J.; Concha, M. L.
2009. Philosophical Transactions of the Royal Society B, 364 (1519), 991–1003. doi:10.1098/rstb.2008.0260
